All ENIGMA protocols are now linked on the ENIGMA GitHub page page. Please check here for the latest amendments and updates!
This page is intended to help guide you key steps for analyzing diffusion MRI while going through the standardized protocols for ENIGMA-DTI.
Please note that these protocols are subject to improvements as we get more feedback, so please check often for the latest!
If you find these protocols useful in your research, please provide a link to the ENIGMA website in your work: www.enigma.ini.usc.edu and tell us what you think! ~ The ENIGMA-DTI Support Team~
We have a network of DTI experts willing to help with any processing questions along the way. email support.enigmaDTI@ini.usc.edu with any DTI related questions!
The basic process is broken down in to 3 main categories, each with its own added set of quality control protocols:
DOWNLOAD OUR PROTOCOLS ON NITRC
Preprocessing ENIGMA-DTI Processing GWAS Analysis
Preprocessing
ENIGMA-DTI Software tools for Preprocessing Quality Control:
- ENIGMA-DTI_QC_FA_V1: Check vector directions (Do gradients in V1 align appropriately along FA image?).
Useful Tools/Tutorials/Pages:
- dcm2nii: DICOM to NIFTI conversion
- FSL: Many image processing tools used throughout ENIGMA-DTI protocols
- CFARI: Many image processing tools useful for DTI/Q-ball processing
- MedINRIA Various diffusion processing tools
- diffusion-imaging.com lots of practical information on DTI!
- cabiatl DTI tutorial
- LPCA denoising DWI denoising
- email me to add your favorite tools here.
We will therefore not require a specific protocol to be followed with any particular software, as long as the appropriate steps are performed. This will allow maximal integration with current pipelines and ensure optimal processing per site if available, and allow sites to:
- process data efficiently with respect to acquisition parameters (eg., do not spend time on HARDI-specific algorithms if you only have 6/12 directions collected)
- take advantage of your scanning protocols
- if you know you can calculate FA more robustly using one of many alternate methods, go for it!
- maximize the quality for your scans (denoising/ removing artifacts etc.)
- keep things in line with current/future projects, and non ENIGMA-related investigations you are working on.
If you have FA measures maps calculated and registered already, we can work with you to include them into the Pipeline rather than to re-run everything from the start. Therefore, if you have discovered workflows and methods that fit your data well to best improve SNR, this would be ideal.
If you have already processed your data, please email support.enigmaDTI@ini.usc.edu to let us know your processing workflow. Also if you would like to update this page with any particulars for your methods, please let us know and we would be happy to work in additional options.
For those that have yet to process DTI data, various suggestions are outlined here. A basic series of steps are as follows: NOTE: most of this can be done in multiple ways depending on your data. please do not hesitate to contact us for support .
- Convert DICOM images to DWI set and T1-weighted set and other data acquired.
- Determine how your DWI set(s) are organized
- How many many acquisitions do you have? Multiple acquisitions can be merged for optimal signal-to-noise ratio.
- How many b0s do you have and where are they with respect to the full series? (Often the b0 image(s) is/are the first volumes in the DWI set)
- If you have multiple b0, were they acquired with the same encoding gradient? If not, slight variations in processing will be needed.
- Correct for Eddy Current distortions, movement using affine registration.
- A convenient option for this is FSL's "eddy_correct" command.
- You can use this script to rotate your bvec files (gradient directions) after using FSL's "eddy_correct" command. (You can use the output of the file from now on and to create your FA images (e.g. as input to dtifit) )
- Create a mask for your data.
- FSL's bet2 offers a solution that is quite robust for many datasets.
- Correct for EPI induced susceptibility artifacts -- this is particularly an issue at higher magnetic fields.
- If you have two opposing b0s and a sufficient amount of diffusion directions obtained, you may use FSL's TOPUPand EDDY for distortion correction.
- If a fieldmap has been collected along with your data, FSL's FUGUE tool may help compensate for the distortions.
- Alternatively, a subject's DWI images can be adjusted through high dimensional warping of the b0 to a high-resolution structural (T1- or T2- weighted) image of the same subject not acquired using EPI. This requires multiple steps:
- Make sure skull-stripping has been performed on both b0 and T1-weighted scans.
- Make sure T1-weighted scans have undergone inhomogeneity (NU) correction.
- Make sure T1-weighted scans and the DWI are aligned!! Check for L/R flipping!!
- Linear registration of b0 of DWI and T1-weighted reference together. **Due to differences in resolution and further registrations needed throughout the workflow we recommend initially aligning the the T1-weighted scans to ICBM space (which is the space of the ENIGMA-DTI template), then using a linear registration (with NO sheering parameters) to align your b0 maps to their respective T1-weighted scans in ICBM space**
- If using FSL's flirt for linear registration, sheering can be avoided by manually setting the degrees of freedom (default 12) to 9 (flirt -in b0.nii.gz -out b02T1w.nii.gz -df 9 -ref T1w.nii.gz)
- Once images are in the same space and linearly alight (visually check this!), you can perform non-linear registrations to remove the distortion from the b0.
- Some possible tools include ANTS , DTI-TK, or BrainSuite.
- The deformation fields from the warping should then be applied to all volumes in the DWI.
- Calculate tensors (this can be done in multiple ways depending on your data).
- Most tools will also output FA, MD, and eigenvalue and vector maps simultaneously.
- FSL's 'dtifit' command is an acceptable and convenient option. It uses least-square fitting to determine the tensor and will output FA and V1 (primary eigenvector) needed for future analyses.
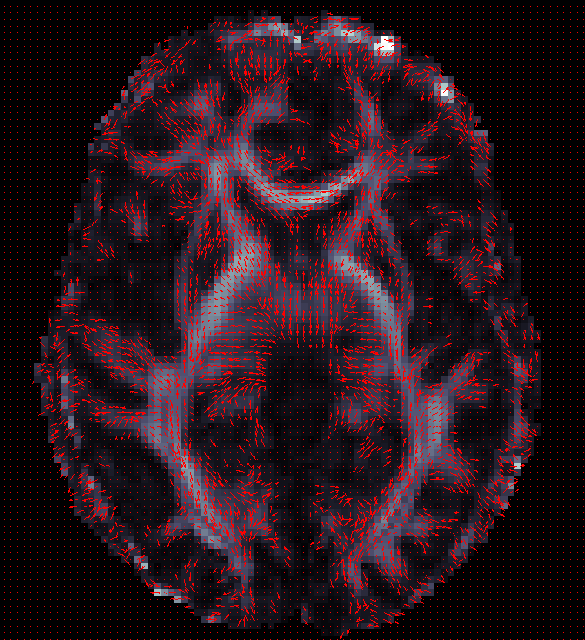
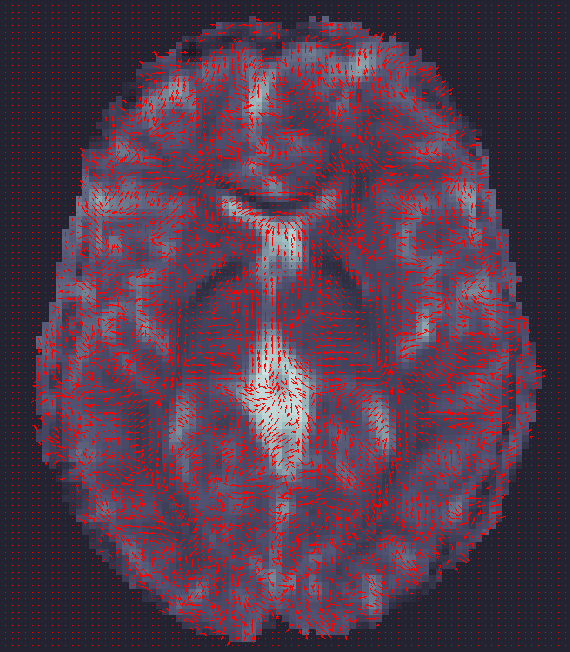
ENIGMA-DTI Skeletonization
Protocols
- ENIGMA-TBSS protocol (PDF format)
- ENIGMA-TBSS protocol (Word format)
- Once DTI data have been pre-processed, use the protocol above to map your images onto the ENIGMA-DTI FA template and project the skeleton.
- Do not forget to QC your images after FA maps have been registered! Misaligned images can cause problems with the overall GWAS and are difficult to detect once skeletonized.
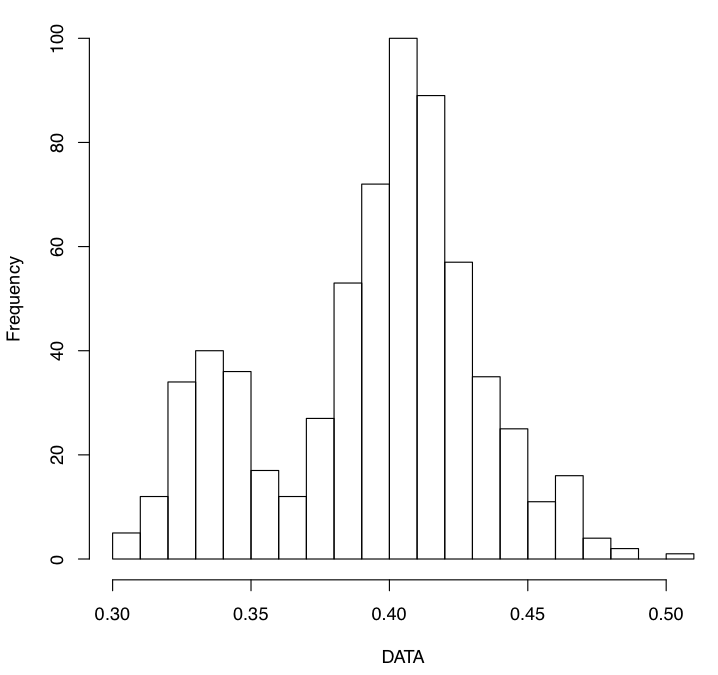
- Use the script here to check the values of the projection distances . Make sure there are no outliers!
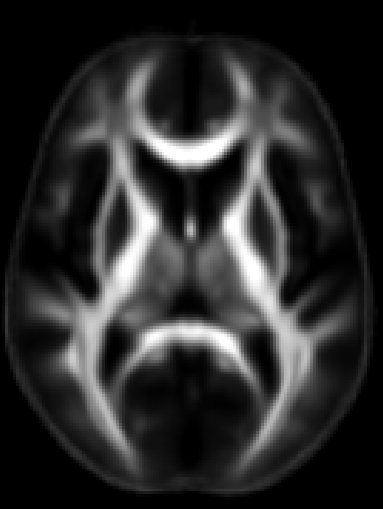
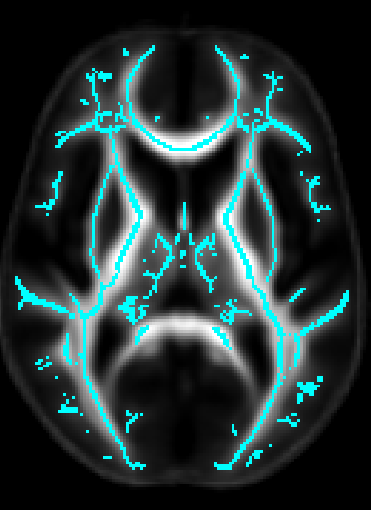
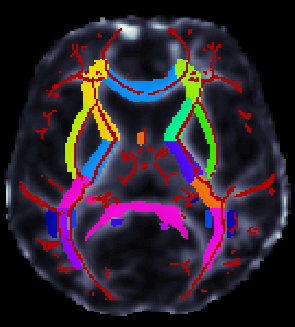
ROI extraction from FA images
- ENIGMA-ROI extraction protocol (PDF format)
- ENIGMA-ROI extraction protocol (Word format)
- Use the above link to extract regions of interest from the skeletons and calculate the average FA within them.
**** An important note **** in the analysis is that the ROI labeled "IFO" is actually different that the same ROI under the most current FSL JHU Atlas label. This was different in the older JHU atlas used to create the template and make the protocols. Please note this will not play a role in ENIGMA-DTI GWAS as we will not be using this ROI for GWAS (it is very small) but it may be considered for disorder studies.
If groups choose to use these measures for their own analysis, please be advised that this should be uncinate according to the current atlas. To avoid confusion, we will NOT switch the labels back, but we will keep this warning so you can carefully examine your data.
ENIGMA-DTI Software tools for ENIGMA processing Quality Control:
- ENIGMA FA/Skel check: Check FA Registrations and make sure skeletons are correct.
- ENIGMA DTI SummaryStats protocol: Check the distribution of your ROIs and look for outliers.
More DTI tools:
Skeletonization of diffusivity measures including mean, axial and radial diffusivity (MD, L1, RD)
If you would like to skeletonize non-FA diffusivity measures including mean (MD), axial (AD/L1) and/or radial diffusivity (RD) and then extract relevant ROI information from them according to the ENIGMA-DTI template, please follow the instructions provided at the below link:
___________________________________________________________________________________________________
Analysis
| GWAS |
We will be performing 2 large scale GWAS:
- for all healthy controls and patient populations
- for all healthy controls only
- a 3rd GWAS for all patient populations without controls will be performed as a possible follow-up analysis
GWAS will be performed for the average FA value in the following regions of interest:
For samples without related individuals, associations will be run using Mach.
To account for relatedness within family-based datasets, we recommend and provide instructions for running merlin-offline a part of the Merlin package here .
All related and necessary software will be downloaded as part of the scripts, so no need to download them from here now!
Please follow the following link to access scripts for running the DTI-GWAS:
Easy to use SCRIPTS
Use this script after skeletonizing your FA images to check the mean and max projection distances to the skeleton:
#!/bin/sh
# Emma Sprooten for ENIGMA-DTI
# run in a new directory eg. Proj_Dist/
# create a text file containing paths to your masked FA maps
# output in Proj_Dist.txt
# make sure you have FSL5!!!
###### USER INPUTS ###############
## insert main folder where you ran TBSS
## just above "stats/" and "FA/"
maindir="/enigmaDTI/TBSS/run_tbss/"
list=`find $maindir -wholename "*/FA/*_masked_FA.nii.gz"`
## insert full path to mean_FA, skeleton mask and distance map
## based on ENIGMA-DTI protocol this should be:
mean_FA="/enigmaDTI/TBSS/ENIGMA_targets/mean_FA_MyMasked.nii.gz"
mask="/enigmaDTI/TBSS/ENIGMA_targets/mean_FA_skeleton_MyMasked.nii.gz"
dst_map="/enigmaDTI/TBSS/ENIGMA_targets/enigma_skeleton_mask_dst.nii.gz"
##############
### from here it should be working without further adjustments
rm Proj_Dist.txt
echo "ID" "Mean_Squared" "Max_Squared" >> Proj_Dist.txt
## for each FA map
for FAmap in ${list}
do
base=`echo $FAmap | awk 'BEGIN {FS="/"}; {print $NF}' | awk 'BEGIN {FS="_"}; {print $1}'`
dst_out="dst_vals_"$base""
# get Proj Dist images
tbss_skeleton -d -i $mean_FA -p 0.2 $dst_map $FSLDIR/data/standard/LowerCingulum_1mm $FAmap $dst_out
#X direction
Xout=""squared_X_"$base"
file=""$dst_out"_search_X.nii.gz"
fslmaths $file -mul $file $Xout
#Y direction
Yout=""squared_Y_"$base"
file=""$dst_out"_search_Y.nii.gz"
fslmaths $file -mul $file $Yout
#Z direction
Zout=""squared_Z_"$base"
file=""$dst_out"_search_Z.nii.gz"
fslmaths $file -mul $file $Zout
#Overall displacement
Tout="Total_ProjDist_"$base""
fslmaths $Xout -add $Yout -add $Zout $Tout
# store extracted distances
mean=`fslstats -t $Tout -k $mask -m`
max=`fslstats -t $Tout -R | awk '{print $2}'`
echo "$base $mean $max" >> Proj_Dist.txt
# remove X Y Z images
## comment out for debugging
rm ./dst_vals_*.nii.gz
rm ./squared_*.nii.gz
echo "file $Tout done"
done